THE USADEL LAB
Cell walls, metabolism and bioinformatics
Home
The Usadel group turns plant omics big data into insight for crop improvement and a sustainable bioeconomy. We use comparative genomics and pangenomics of crops and their wild relatives – in particular Solanaceae (tomato, potato, eggplant), Brassicaceae (canola, camelina), berry crops such as blackcurrant and strawberry, tea, and emerging protein crops like faba bean – to uncover genes and mechanisms that drive abiotic stress resilience and the accumulation of valuable or health-promoting compounds.
Our research combines state-of-the-art long-read sequencing, genome assembly and phasing with integrative analyses of genomes, transcriptomes and metabolomes. By building high-quality reference genomes and pangenomes and linking them to detailed phenotyping, we can dissect structural variation and complex traits such as drought and disease tolerance, flavour and nutritional quality.
A second core pillar of the lab is automated genome interpretation. We develop widely used tools and ontologies – including MapMan, the Mercator functional annotation pipeline and the deep-learning gene finder Helixer, together with plant genome platforms such as PlaBiPD and PubPlant – to connect gene sequences to functions, pathways and quantitative trait loci at scale. In parallel, we contribute to national and international FAIR data initiatives (e.g. NFDI4Plants/DataPLANT), helping to ensure that plant research data remain findable, accessible and reusable for the community.
Latest news:
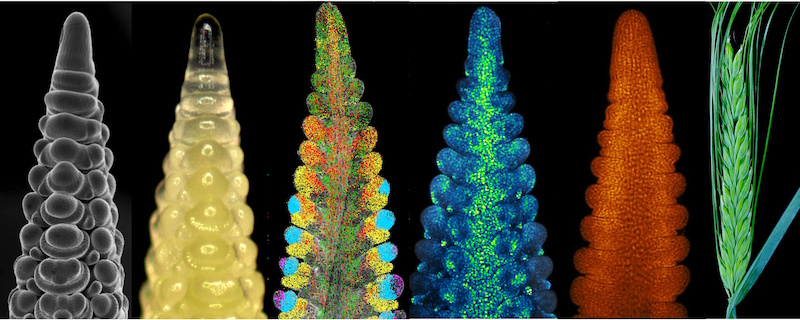
Going spatial with CEPLAS
General | Jan 14, 2026
By integrating deep single-cell RNA sequencing with spatial transcriptomics from...

Structural gene annotation
General | Nov 26, 2025
Helixer a tool for structural annotation has been published in Nature Methods. T

Nanopore Day at HHU
General | Nov 19, 2025
We were hosting the nanopore day "The Power of Oxford Nanopore Technologies in Plant...

Eggplant pangenome assembled and analyzed
General | Nov 18, 2025
With our dear friends we managed to unravell the eggplant pangenome